Scalable PDF Document Processing with DataChain and Unstructured.io
The open source versions of DataChain and Unstructured.io can work together to scale PDF document processing. In this tutorial you will learn the steps to accomplish this including, how to create and save the DataChain, defining the UDF, and how DataChain versioning works.

Most organizations keep a large source of information in the form of various internal documents, call transcripts and other unstructured data. These data contain a lot of useful insights about customers, employees or the inner workings of the company. However, they remain largely untapped by data teams due to the difficulty of dealing with large quantities of data in unstructured formats.
Today, we will see how you can process a collection of documents in less than 70 lines of code, extract and parse text from them and create vector embeddings useful for downstream tasks (e.g. for RAG or as ML features). This approach is also scalable and you will benefit from easy versioning of the final datasets.
Approach and tools
We will work with a publicly available Google Storage bucket which contains a collection of Neurips conference papers (representing our internal company documents).
For data processing we will use the Unstructured.io Python library which contains a lot of useful functionality for unstructured data processing. They also offer an API to handle processing on their compute and offer some extra options and features on top of their FOSS offering. We will be using the API for partitioning to take advantage of advanced partitioning features.
With unstructured we will:
-
Easily ingest and partition each document using the Unstructured API
-
Clean the partitions
-
Create vector embeddings from the partitions
We will also use DataChain, which is an open-source Python data-frame library that helps ML and AI engineers to build a metadata layer on top of unstructured files. DataChain enables out-of-memory storage and processing with a Pythonic dataframe-like API that combines SQL-type operations with GPU/CPU acceleration and seamless scalability while also versioning and persisting datasets for reproducibility.
With DataChain, we will:
-
Easily search and filter our data container to only load the documents we need
-
Scale up the document processing with unstructured.io to the level of our entire document collection
-
Save the results as versioned datasets in tabular format, ready for processing downstream
Both libraries can be easily installed with pip.
pip install "unstructured[pdf,embed-huggingface]" datachain(In this example we are using unstructured version 0.15.7)
For the Unstructured API, we will also install the python-dotenv library to
load the API key and URL. To get the API key, you can follow this guide and get
a free API key for up to 1000 document pages per month.
We then need to save a .env file with the following content in the location we
will be running our code from:
UNSTRUCTUREDAPI_KEY=<YOUR_APIKEY>
UNSTRUCTUREDAPIURL=https://api.unstructured.io/general/v0/generalFull working code
Here you can have a look at the full code used in our example which you can run
(and we will unpack in a second) This code will load our document collection
with DataChain and create a DataChain UDF (user-defined function) process_pdf
which will load, partition and clean the text and create vector embeddings using
unstructured. It then saves (and automatically versions) the resulting dataset
containing the embeddings, cleaned human-readable text and a reference key for
all of the original documents.
import os
from collections.abc import Iterator
from dotenv import load_dotenv
from datachain import DataChain, C, File, DataModel
from unstructured.partition.api import partition_via_api
from unstructured.cleaners.core import clean
from unstructured.cleaners.core import replace_unicode_quotes
from unstructured.cleaners.core import group_broken_paragraphs
from unstructured.embed.huggingface import (
HuggingFaceEmbeddingConfig,
HuggingFaceEmbeddingEncoder,
)
load_dotenv()
# Define the output as a DataModel class
class Chunk(DataModel):
key: str
text: str
embeddings: list[float]
# Define embedding encoder
embedding_encoder = HuggingFaceEmbeddingEncoder(config=HuggingFaceEmbeddingConfig())
# Use signatures to define UDF input/output (these can be pydantic model or regular Python types)
def process_pdf(file: File) -> Iterator[Chunk]:
# Ingest the file
with file.open() as f:
chunks = partition_via_api(
file=f,
metadata_filename=file.path,
api_key=os.getenv("UNSTRUCTURED_API_KEY"),
api_url=os.getenv("UNSTRUCTURED_API_URL"),
content_type="pdf",
strategy="fast",
chunking_strategy="by_title",
)
# Clean the chunks and add new columns
for chunk in chunks:
chunk.apply(
lambda text: clean(
text, bullets=True, extra_whitespace=True, trailing_punctuation=True
)
)
chunk.apply(replace_unicode_quotes)
chunk.apply(group_broken_paragraphs)
# create embeddings
chunks_embedded = embedding_encoder.embed_documents(chunks)
# Add new rows to DataChain
for chunk in chunks_embedded:
yield Chunk(
key=file.path,
text=chunk.text,
embeddings=chunk.embeddings,
)
dc = (
DataChain.from_storage("gs://datachain-demo/neurips")
.settings(parallel=-1)
.filter(C.file.path.glob("*/1987/*.pdf"))
.gen(document=process_pdf)
)
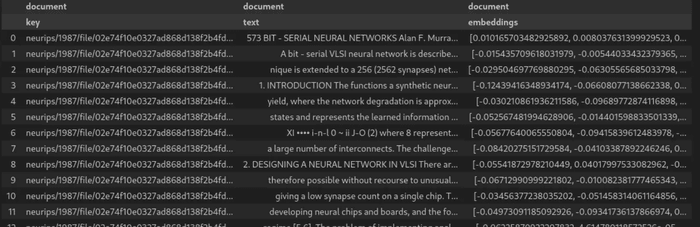
dc.save("embedded-documents")
DataChain.from_dataset("embedded-documents").show()The resulting dataset will look like in the image below:
How it works
Creating and saving the DataChain
The following few lines of code are all that we need to load and select the
right data in from our storage and to process them with unstructured.
dc = (
DataChain.from_storage("gs://datachain-demo/neurips")
.settings(parallel=-1)
.filter(C.file.path.glob("*/1987/*.pdf"))
.gen(document=process_pdf)
)Let’s unpack:
The from_storage and filter methods allow us to ingest the data from a
bucket/storage container. Since DataChain uses lazy evaluation, no other files
than those specified by the filter will be loaded, speeding up the process
considerably. Setting parallel to -1 tells DataChain to make use of all
CPUs/cores on the current machine, speeding things up once again.
This will create a DataChain metadata table containing all the information needed to process our PDF files without actually having to copy the files themselves or load them all to memory. Since DataChain operates on a metadata level, it can scale up billions of files without us having to worry about memory overflows.
The gen method allows us to modify the DataChain table and create new rows
(potentially more than one per original table row) using DataChain UDF
functions. Here, the UDF process_pdf does all the individual PDF processing
with unstructured.
Finally, we save the table as a dataset by calling
dc.save("embedded-documents")This will persist the table and version it (each time we call this command a new version is created automatically). We can then load and display it by the following command, optionally specifying the dataset version (more on that later).
DataChain.from_dataset("embeddings").show()All that’s missing is the DataChain UDF definition, so let’s see how we do that.
Defining the UDF
The process_pdf UDF will take the original DataChain table and produce an
output with embeddings of processed document chunks. We also want to keep the
original text of each processed document chunk and a key by which we can
link each chunk back to the original full document.
We first specify what we actually want to receive on the output of our UDF by
defining the DataModel-based Chunk class and defining the output column
types:
# Define the output as a DataModel class
class Chunk(DataModel):
key: str
text: str
embeddings: list[float]Now we are ready to define the process_pdf function itself. We use Python
signatures to specify the input and output. Here, File is a class used by
DataChain to refer to the original file - a PDF document in our case. On the
output we use Iterator since our function will produce multiple chunks (and so
multiple rows in our DataChain table) per original file.
def process_pdf(file: File) -> Iterator[Chunk]:The rest of the function definition as well as the definition
of embedding_encoder specifies how unstructured is used to process each
individual PDF file. We are using
partition_via_api
to take advantage of the more advanced "by_similarity" chunking strategy only
offered in the API. If we are not interested in that, it is also possible to do
the same using only the free open source unstructured library by replacing
partition_via_api in the script above as follows:
from unstructured.partition.pdf import partition_pdf
...
chunks = partition_pdf(file=f, chunking_strategy="by_title", strategy="fast")For more detail on data processing with unstructured you can check out the
tutorial in the
unstructured documentation.
Here, we will just note that if some of our documents were scanned images or
contained tables or other more complicated elements we would replace "fast" in
the strategy parameter of partition_pdf or partition_via_api with
"hi-res". Then unstructured will be able to process that kind of data.
Finally, we want the UDF to produce new rows in our DataChain table and so we
have it return the Chunk objects we specified above. Here, we use yield
instead of return as each PDF file produces several Chunk objects.
# Add new rows to DataChain
for chunk in chunks_embedded:
yield Chunk(
key=file.path,
text=chunk.text,
embeddings=chunk.embeddings,
)DataChain versioning and additional SaaS features
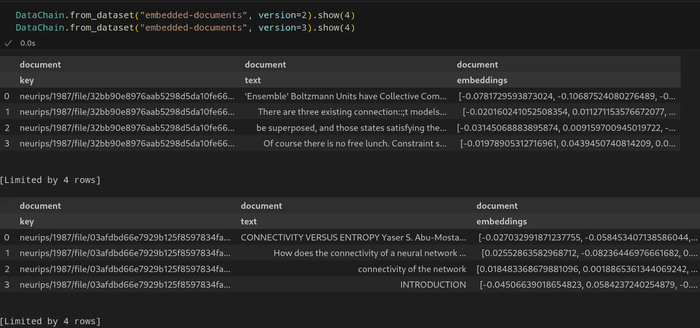
Now, we have a versioned dataset with a new version automatically created by DataChain whenever we run the script above. By default, versions are numbered and we can always recall a particular version by specifying it as in this call:
DataChain.from_dataset("embedded-documents", version=<version number>).show()
In the SaaS version of DataChain we also provide lineage tracking for datasets, more robust dataset versioning and auditability, a managed on-demand compute including with GPU clusters to really scale up your workloads and a graphical user interface among other features.
DataChain SaaS is currently available as a private preview in our Studio platform right now, feel free to ask us about that at [email protected]!
Summary
We have processed our collection of documents to create a dataset of embeddings in a scalable fashion. We could now proceed with optimizing the workflow, choosing the best way to pre-process our data by trying different parsing and data cleaning strategies, different embedding etc.
Unstructured provides many tools for such fine-tuning and dataset versioning with DataChain is a useful tool to compare the different iterations. With the DataChain SaaS it is also very easy to scale this up further with managed cluster compute and robust lineage and versioning capabilities. We will look at this in more detail next time!
You can also find this content on video here: